The Curriculum Unit
The curriculum unit is arranged in three parts. Part 1 presents a review/overview of molecular genetics. There is a specific focus on how information encoded in deoxyribonucleic acid (DNA) results in protein products and how the mutations of various kinds can interfere with those processes. Part 2 introduces gene therapy and the mechanisms thereof. A general overview is presented, but there is also a focus on specific methods being developed and employed by researchers as potential treatments for DMD. Part 3 asks students to employ their scientific learning to engage with some of the ethical and moral topics that are raised by gene therapy research and the advances thereof.
Part 1 - Overview of Molecular Genetics
It is widely understood that DNA is a biomolecule that contains the blueprint for any living organism. Within the base pairs of the double-helical structures housed in a single nucleus is all the information needed to build the entire organism or any part therein. In short, DNA is the molecular basis for heredity. As such, DNA must both preserve the genetic information and facilitate the expression of the genetic information.
The information stored in DNA is the result of the arrangements of its four chemical bases: adenine (A), guanine (G), cytosine (C), and thymine (T). Akin to the manner in the which letters of the alphabet can be arranged to form words and sentences with an endless variety of meanings, so too can these A, G, C, and T bases of DNA be ordered to hold an array of genetic information. This framing device of using letters and words to construct complex sentences with clear meanings is the through line of the unit. In the end, and at points throughout the unit, students construct examples and models to show foundational concepts underpinning molecular biology and gene therapy.
According to the central dogma of genetics, the pattern of information occurring most frequently in cells is as follows: information within DNA is preserved when the molecule is duplicated through the process of replication and in order for the genetic information to be expressed, DNA is transcribed into RNA, then RNA is translated into protein. DNA molecules are arranged in a double helix structure in which bases pair complementarily with each other, A with T and C with G. In this way, each strand of the double helix can serve as a template to reproduce the entire molecule (replication). Replication is essential because in most cell divisions each new cell must have a complete copy of the DNA contained in the original cell. To facilitate gene expression, a DNA sequence on one strand is used as a template to produce an RNA sequence (transcription), which in turn is to be used as guide for assembling protein (translation) (Figure 2).

DNA is the molecular basis for heredity because the order of its bases, the gene sequence, determines the protein products that will be produced. Genes are the specific segments of DNA, a specific set of instructions, that encode particular protein products. Genes can range in size from as small as a few hundred base pairs long to as large a million or more base pairs. According to the Human Genome Project, the human genome contains approximately 3 billion base pairs divided among 23 pairs of chromosomes. Within these base pairs is encoded information for approximately 25,000 genes (Griffiths, et al. 2005).
With billions of base pairs and tens of thousands of genes, it is necessary to have a comprehensive coding system to organize genetic information. To recall the earlier analogy of the alphabet and sentence construction, it is necessary to understand how letters are grouped to form words and how the words of a sentence are read in sequence across a page to convey complete ideas. In an analogous manner, it is necessary to know that the genetic code is a specific (unambiguous), universal, non-overlapping, degenerate (redundant), non-punctuated code in which groups of three bases (codons) code for the amino acids used to construct protein products (Griffiths, et al. 2005).
To begin, there are 20 different amino acids commonly found in cellular proteins. These in a way are the words that form sentences (proteins).
When a strand of messenger RNA is read end to end, there is only one base at each position. As there are only four different bases in RNA (A, U, G, and C), amino acids cannot be represented by a single letter (base); otherwise the size of the vocabulary, four, would be far too small to accommodate the 20 words (amino acids), needed to form fully coherent, complex, and meaningful sentences (functional proteins). If pairs of letters (AA, AU, GC, etc.) were the basis of word formation, the vocabulary would be larger (42 = 16), but still insufficient. With triplet codons (AAA, CGC, AUG, etc.), 64 (43) distinct word combinations are possible, more than enough for 20 amino acids. Experiments by Francis Crick and other researchers confirm that the codon is indeed exactly three bases long (Griffiths, et al. 2005). The genetic code is said to be degenerate (redundant) because some amino acids are in encoded by multiple codons, however it is also specific (unambiguous) in that no codon codes for multiple amino acids (Figure 2). It is also important to note that the code is non-punctuated; once reading of the letters (bases) is initiated, each letter is read until a stop codon is reached – no commas, no spaces. Crucially, adjacent words (amino acids) do not share letters (bases); the code is said to have non-overlapping reading frames.
By applying these rules of genetic sentence construction, a DNA sequence such as ATGACGGATTAG, is transcribed into the messenger RNA transcript AUGACGGAUUAG and then translated into the polypeptide sequence Met-Thr-Asp (Figure 2).
Mutations
The framing device of sentence construction is helpful beyond just understanding how a DNA code reliably determines how proteins are constructed. It is also useful for understanding the consequences of errors in those processes, mutations in particular. Gene mutations are lasting changes in the DNA sequences that code for a protein. The causes of mutations are many and include random errors in the DNA replication process. The source of a mutation can involve a single base, a few bases, of even large segments of DNA including entire chromosomes. For the purposes of this curriculum unit, it is important to understand several types of mutations and even more so to grasp the potential consequences for the protein product of the associated gene.
Missense mutation – Missense mutations occur when a single base pair substitution occurs such that one amino acid in the peptide sequence is replaced with a different amino acid (Figure 3). The severity of the consequences will differ depending on the role of the particular protein, the location of the mutation, and the differences between the amino acids; a relatively benign missense mutation might slightly affect the shape and folding of protein without resulting in significant losses or gains of function. In other cases, a single amino acid change in a critical domain of the protein might result in a very diminished protein product and/or critical loss of function. The mutation that is associated with sickle cell anemia, for example, is caused by a single base pair substitution in which a GAG codon in the gene for hemoglobin is changed to GTG resulting in an amino acid valine where there should be a glutamic acid (Kumar, Fausto and Abbas 2004). The consequence of this minor change in structure is a significant shift functional capability.
Nonsense mutation – Nonsense mutations occur when a single base pair substitution occurs such that one amino acid in the peptide sequence is replaced with a stop codon; a signal for the translation apparatus to stop building the protein (Figure 3). Like the single base pair missense mutations, the severity of the consequences of a nonsense mutation depends on the particular protein and the location of the mutation. In general, though, the closer a nonsense mutation is to the beginning of a gene, the more likely it is to have significant effects as it is more likely to disrupt critical domains downstream from the substitution. A sufficiently truncated protein might retain no function at all. Some of the most severe forms of DMD, for example, are the result of nonsense mutations early in the sequence of the dystrophin gene (Kumar, Fausto and Abbas 2004).
Insertion and Deletion Mutations– Insertion mutations add one or more bases somewhere in the sequence of the gene, whereas, deletion mutations remove one or more bases from the sequence of the gene (Figure 3). Very large deletions might even remove the whole gene. The potential consequences range depending on the nature of the particular sequences added or removed as well to/from where they are added or removed. These consequences are discussed further following the description of frameshift mutations (Kumar, Fausto and Abbas 2004).
Duplication Mutations – Duplication mutations are abnormal repetitions of one or more base pairs. As with other mutations, severity of consequences varies. These consequences are discussed further following the description of frameshift mutations (Kumar, Fausto and Abbas 2004).
Frameshift Mutations – Frameshift mutations occur when the insertions, deletions, duplications, or similar mutagenic events disrupt the reading frame of the gene sequence (Figure 3). Recall that according to the sentence construction analogy established previously, the genetic code is non-overlapping and non-punctuated. Also, amino acids are constructed based on codons defined by base triplets. As such, for mutations that change the number of bases in the gene sequence, mutations that add or remove bases in multiples of three and occur between instead of within codons are likely to be less consequential than others. For frameshift mutations, the protein product is often nonfunctional because all downstream codons are likely to be altered (Kumar, Fausto and Abbas 2004).

Along with the previous descriptions and the images in Figure 3, the sentence construction framework is used to emphasize how changes in gene sequences can have a range of consequences on the protein product associated with a gene.
To illustrate the utility of this framework, consider a sentence comprising a sequence of three-letter words. As described previously, the letters in the sentences represent individual DNA bases in the gene and the words represent triplet codons coding for amino acids. The complete sentence represents the protein. The meaning conveyed by the sentence is also essential for the utility of the sentence construction framework in illustrating and understanding the consequences of mutation. The intended meaning of the sentence represents the function of the protein. In this construct, a variety of mutations can be simulated by adding, removing, or changing one or more letters to/from/in a sentence and assessing the extent to which the originally intended meaning of the sentence is affected. The extent to which the originally intended meaning of the sentence is distorted is a stand-in for how much of the function of a particular protein is changed after a mutagenic event.
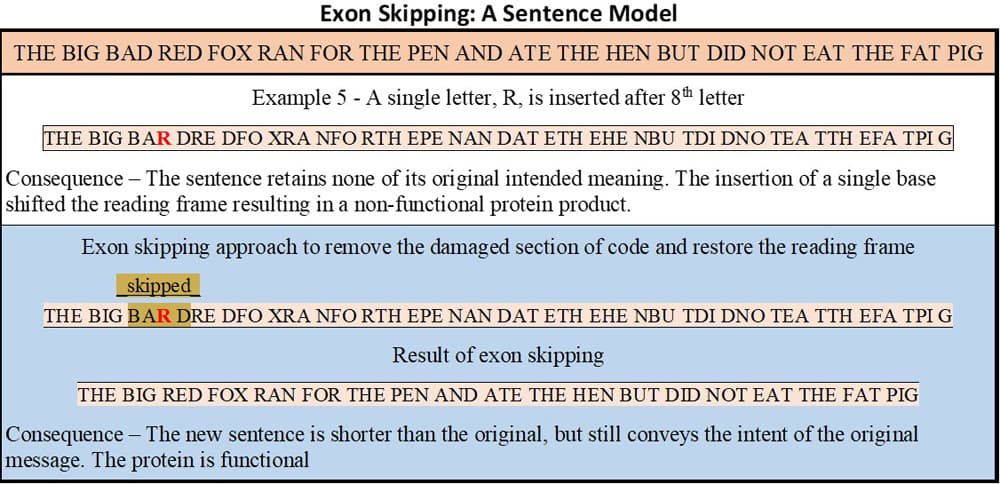
Consider the sentence and the consequences of the example “mutations” shown in figure 4.

Muscular Dystrophy and the DMD Gene
Duchenne muscular dystrophy (DMD) is a neuromuscular disorder affecting roughly one in every 3500-5000 male births worldwide. DMD is characterized by progressive skeletal muscle degeneration and weakness. Early symptoms begin with the legs and pelvis which are associated with the large muscle groups of the legs and gluteal region, but also occur less severely in the arms, neck, and other areas of the body. The first symptoms are usually observed between ages 3 and 5 and often include difficulty standing upright from a laying position and frequent falls. These boys also tend to have trouble climbing stairs. Over time the condition progresses and the motor skill deficits (running, hopping, jumping, etc.) increase. In time, even walking becomes too challenging and many of these boys are confined to wheelchairs by age 12. Beyond this point sleeping and breathing aids also become necessary. By age 20 most DMD patients are characterized by severe breathing difficulties and cardiac disease. Most do not survive beyond the third decade of life due to respiratory or cardiac complications (Blake, et al. 2002).
In this section of the unit, students’ comprehension of the primary functions associated with the muscular system (described previously) is reinforced through analysis of still images and videos made of DMD patients from around the world. Students analyze patient histories of people who lived or are living of with DMD. Students also watch and read, interviews and accounts by DMD patients and their loved ones and caregivers. Where the opportunity is possible, students conduct interviews with DMD patients, their families, and physicians who treat DMD patients. Gaining familiarity with a condition that highlights, in an almost visceral manor, the functional consequences of the living with less muscle, underscores for students the roles that their own muscles play, even when they are not thinking about them.
According to the 3 P’s, the functional consequences associated with DMD can be sourced to changes in structure. At the level of muscle tissue, students emphasize the connection between the small boys’ small frames and their diminished abilities to move. Students zoom in further to the level of the cell and draw connections between the mutated dystrophin gene and the reduced ability of muscle cells to remain stable during contraction-relaxation cycles.
DMD is caused by an absence of normal quantities of dystrophin, a cytoskeletal protein that stabilizes the plasma membrane of muscle fibers helping to keep muscle cells intact. Loss-of-function mutations in the DMD gene coding for dystrophin diminish the muscles’ ability to withstand physical stresses owing to lengthening and shortening associated with repeated muscle contractions (Kendall, et al. 2012).
DMD largely affects boys because it is inherited in an X-linked recessive fashion due to the location of the DMD gene on the short arm of the X-chromosome. At 2.4 Mb, the DMD gene is among the largest in the human genome. Even after introns (nucleotide sequences that are not included in the final RNA product to be translated) are removed from the precursor messenger RNA (pre-mRNA) transcript, the mature DMD mRNA transcript produced by splicing its 79 exons (regions coding the final protein product) is still 14 kb in length. The sheer size of this gene and its transcript make the DMD gene more susceptible to mutations, but research also shows that certain regions of the DMD are mutation hotspots (nucleotide sequences with a higher concentration of mutations (Rodino-Klapac, et al. 2007). These mutations exist in the varieties previously described and are associated with the ranges of consequences modeled by the previous sentence construction analogies.
Given these underlying genetic elements of DMD, the availability of locus-specific databases of DMD mutations is invaluable for research, diagnosis, and clinical care. Advances in gene sequencing technologies have helped in developing just such databases that can aggregate data from groups of DMD patients. The unit introduces students to such data from the United Dystrophinopathy Project (a multicenter research consortium) and the TREAT-NMD DMD Global database. Analyses of these kinds of data of known DMD gene mutations find that deletions comprise the majority, but duplications and point mutations are also known (Bladen, et al. 2015).
As modeled in the sentence construction analogy, the characteristics and locations of specific mutations affect the severity of functional consequences. The range of consequences associated with mutations in the DMD gene includes but is not limited patients who endure occasional bouts of cramping and myalgia, patients who live with a related but milder form of muscular dystrophy called Becker’s muscular dystrophy (BMD), and patients with rapidly progressing DMD (Bladen, et al. 2015). The availability of mutation databases, gene maps, and the DystrophySNPs resource developed by the University of Utah Genome Depot allows students to make direct connections between changes in the DMD gene sequences and consequences in the dystrophin protein. Students’ understanding of types of mutations allows them to explore the information in these data sets, then predict and describe consequences likely to occur in the protein. Students are also able to provide reasonable explanations to support the assertions they make. The sentence construction frame work helps the students develop their own understanding and then provides a useful way to communicate meaningful scientific information to science and non-science colleagues alike.
The example of DMD is used in the unit because it so clearly delineates the connection between a change in structure and a corresponding dysfunction. The exploration of gene therapy later in the unit is to emphasize the idea that even complex, cutting edge technological advances are rooted in the central idea that solutions for dysfunctions mollify the effects of abnormal structure.
Part 2 - Mechanisms of Gene Therapy
Whereas part 1 of the curriculum unit was concerned with elucidating the nature of the structural problems that result in the dysfunctions associated with DMD, the focus of part 2 is on how a clear understanding of the abnormal structures can lead to the development of remedies that regain function by restoring structure. This is the purpose of the 3 P’s construction.
The molecular basis for DMD has been known for more than two decades and consequential advancements in the management of the disease have been achieved in the that time. Indeed, DMD patients are diagnosed more readily and have available to them systems of interventions that allow them to live comparatively more robust lives for longer than in years past. Still, the treatment options for DMD patients remain limited. The interventions available are largely palliative, seeking to mitigate the symptoms of the dysfunction rather being able to address the structural root causes of the disease. Corticosteroids, cardiac and pulmonary medications, orthopedic supports and procedures, and several other factors are among the critical components of a thorough interdisciplinary approach to managing DMD. Improved muscular strength, prolonged ambulation, and improved respiratory function can be expected for most DMD patients presently (Bushby, et al. 2017).
Strategies attempted in pursuit of curative rather than merely palliative therapies have included both the transplantation of healthy muscle progenitor cells into DMD patients as well gene-therapy mediated delivery of functional copies of the DMD gene into patients. Success with the former strategy has been limited by “issues of immune rejection and poor systemic delivery and viability of transplanted cells.” That is, the administered cells don’t survive after transplantation, and are not able to contribute to muscle function. Success of the latter strategy has been limited by issues associated with the large, complex structure of the gene (Lim, Maruyama and Yokota 2017).
Gene therapy is an approach to address genetic disorders by targeting the source of the disorder; that is, gene therapy refers to treating a disease condition resulting from an abnormal version of a gene by introducing modified DNA products with the correct gene into the cells of the patient. The most intuitive application of this broad concept is to introduce a functioning gene to correct the effects of a disease-causing mutation (e.g., introducing a functional dystrophin protein where none is present or it is present in insufficient quantities). However, gene therapy might also be implemented to inhibit a cell from producing a damaging product or even to completely destroy a problematic cell. In this sense, the basic concept of gene therapy is simple, but several challenges make implementation on a large scale more difficult; these include but are not limited to developing appropriate vectors to reliably deliver the genetic material to the desired sites, avoiding immune responses, and not disrupting the normal function of non-related genes (Rodino-Klapac, et al. 2007).
As suggested earlier, previous curative strategies for DMD involving gene therapy were unsuccessful at reliably delivering the entire DMD gene into all the necessary tissues in significant part due to its large size. Recent advances in molecular biology techniques have given rise to a gene therapy strategy that does not necessitate that the whole DMD gene be transferred into cells: exon skipping.
In this exon skipping approach, the translational reading frame of a gene is restored using synthetic nucleic acid analogs called antisense oligonucleotides (AOs). These AOs can bind to pre-mRNA sequences and interfere with splicing. Specific AO sequences can be designed to target complementary sequences in pre-mRNA. The cell’s splicing machinery does not incorporate the targeted sequence in the final transcript. In this way, whole exons or even multiple exons can be targeted for exclusion without disrupting the reading frame (Aartsma-Rus and van Ommen 2017).
In the parlance of the sentence construction analogy, rather than having to write a new, error-free version of a long complex sentence, exon skipping simply allows the reader to jump over the portion of the sentence containing the mistake. So long as the reading frame is kept intact, the resulting sentence, though somewhat shorter, may still be able to convey most of the intended meaning of the original sentence.
Figure 5 recalls the previously discussed example 5 (insertion of a single base resulted in a nonsensical protein product) and uses the sentence construction framework to illustrate how exon skipping can fix the effects of certain mutations.
Eteplirsen
In recent months, the US Food and Drug Administration (FDA) granted approval for the first gene therapy drug, using this novel exon skipping strategy, Eteplirsen. The drug uses an AO designed to bind in a complementary way to—and thus exclude—exon 51 from the final DMD gene transcript. Skipping exon 51 addresses structural problems associated with the mutated gene in approximately 14% of patients with DMD mutations (Lim, Maruyama and Yokota 2017).
In healthy individuals (no mutations), the 2.5-Mb DMD gene is transcribed to produce a 21-kb pre-mRNA transcript: 79 exons separated by introns. The cell’s splicing machinery then removes the intron sequences from the pre-mRNA and combines the remaining exon regions into a 14-kb mature mRNA transcript. This mRNA transcript is used as template to translate a normal 427 kDa protein of 3685 amino acids.
Patients who may be helped by Eteplirsen have a DMD gene with a consequential mutation in or near exon 51 (e.g. a deletion that introduces a premature stop codon or a deletion that shifts the reading frame for the downstream sequence). The RNA produced from these do not result in a functional dystrophin protein.
In Eteplirsen-treated DMD patients the action of the AO binding to a specific portion of the pre-mRNA transcript results in a mature mRNA in which exon 48 is followed immediately by exon 52 and the reading frame is intact. The end result of translation is a shorter, but still functional dystrophin protein (Lim, Maruyama and Yokota 2017).
The FDA approval of Eteplirsen is not without various controversies. This provides opportunities for students to engage with these and other issues from a more informed perspective. In one such issue, some scientists tout the available data from clinical trials and foresee an effective treatment option for certain classes of DMD patients. Other scientists raise questions about the methodologies employed by the researchers and cast a skeptical eye on the reliability of the clinical trial data and on the utility of the drug at this time (Kesselheim and Avorn 2016). Now armed with a clearer, more accessible understanding of the molecular mechanisms associated with the drug’s actions, students can now more meaningfully examine the published data from clinical trials and draw conclusions about the efficacy of the drug and reliability of claims made by the manufacturer. Students construct and communicate informed, evidence-based arguments related to contemporary issues in the scientific community. There is a direct and immediate connection between the students’ learning in class, and significant real-world implications and applications of that learning. In considering the effectiveness and viability of gene therapy as a treatment for DMD, students engage with the question “For whom and under what conditions would you recommend gene therapy as a treatment for DMD?”
Part 3 – Ethical and Moral Issues Raised by Gene Therapy
Although gene therapy seems to be an increasingly-promising treatment option for several conditions, it remains a risky and often controversial technique. In general, gene therapy is only being explored for conditions with no other curative remedies. In the last part of the unit students combine their learning about the mechanisms of gene therapy, their analysis of clinical research and regulatory data along with their capacities both critical and abstract thinking, to engage with ethical and moral issues.
Beyond questions of efficacy and safety, gene therapy is often considered controversial due to associated moral and ethical questions. As with other scientific developments, as the technology improves, questions shift from ‘can we’, to ‘should we’. Among other issues, students consider the following questions presented on the NIH Genetics Home Reference website: How can “good” and “bad” uses of gene therapy be distinguished? Who decides which traits are normal and which constitute a disability or disorder? Will the high costs of gene therapy make it available only to the wealthy? Could the widespread use of gene therapy make society less accepting of people who are different? Should people be allowed to use gene therapy to enhance basic human traits such as height, intelligence, or athletic ability?

Comments: