Classroom Activity 3: Making the Cut with CRISPR
Lesson Overview
In this activity, I want to the students to become the Cas9 protein. At the beginning of class, I plan to provide a short lecture (10-15 minutes) about the Cas9 protein and how it performs. At this time, I would also remind my students of the complementary base pairs of DNA and RNA. Once the lecture concludes, I will ask them to search the entire genetic sequence in the classroom and identify and label as many 5’-NGG-3’ sequences possible with Cas9 sticky notes. Afterwards, I will assign schizophrenia-associated miRNA sequences to teams of students, and ask them to write the complete double-stranded DNA sequences that would match the miRNA. Then they will search the DNA sequence around the classroom for the places where Cas9 would actually create edits. By the end of the lesson, I expect students to be able to describe how Cas9 will specifically target DNA using specific amino acid residues and RNA. The activity could be used as part of a cumulative assessment.
Preparation
Below are five miRNA sequences and their complementary DNA sequences related to schizophrenia (Table 1).29
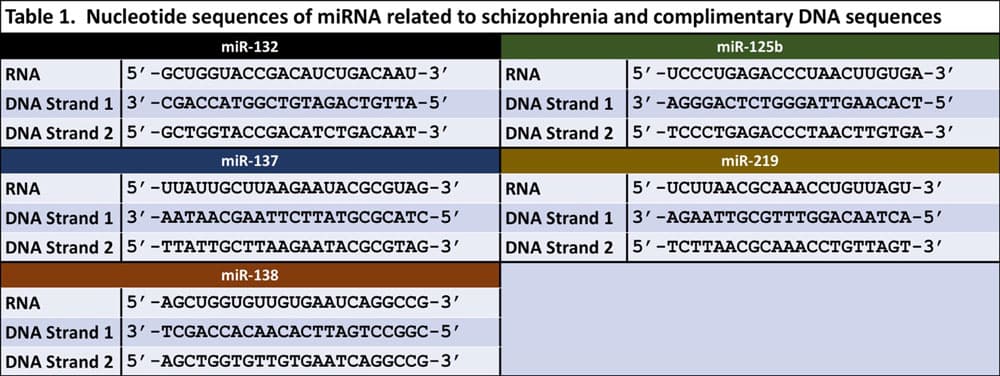
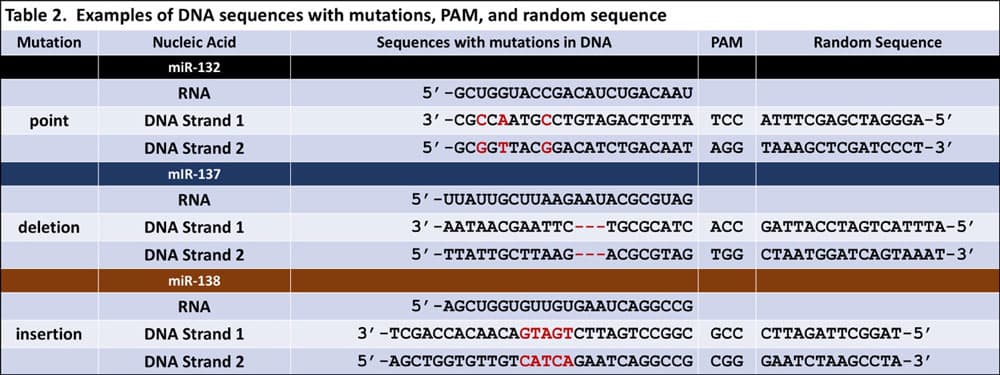
The key to this activity is having enough DNA strands posted around the classroom. To prepare, first generate each of the 5 DNA sequences and introduce a type of genetic mutation into the DNA. Then add the PAM 5’-NGG-3’ to the mutated sequence Finally, add a random nucleotide sequence. Examples can be found in Table 2. Ultimately, you want to create a DNA sandwich with the mutated DNA sequences between two random sequences, knowing that the PAM must be at the 3’ end of the mutated sequence so that it is a designated marker for Cas9 to identify.
Implementation
To start the activity, only give the students with the miRNA sequences. Ask them to derive the DNA associated with the miRNA. To avoid confusion, make sure that the students’ top strand of DNA begins with 3’ and the bottom strand begins with 5’. Focusing on strand 2 as demonstrated in Tables 1 and 2 will help the students find their DNA targets.
Steps to Cutting with Cas9
The following are some basic instructions that you can provide the students.
- Identify the protospacer adjacent motifs (5’-NGG-3’).
- Determine if the four nucleotides (protospacer) to the left of the PAM are complementary to the nucleotides from any of the miRNA sequences. If the they don’t match, then Cas9 will not try to correct the DNA.
- Compare the DNA sequence derived from the miRNA with the DNA sequences provided.
- Determine if the Cas9 protein will correct a mutation, add/delete nucleotides, or create further complications in the DNA.
Once the students have determined the complementary DNA sequences to the miRNA, I will ask them to revisit all of their Cas9 markers placed around the room. Then they will examine the DNA sequence to the left of the Cas9 marker to determine if the Cas9 protein will cut the DNA. If it will not cut the DNA, then students will remove the Cas9 sticky note. If the first four nucleobases (the protospacer) match the DNA, then Cas9 will know to cut the DNA and continue making repairs. Students should continue to read the sequence to determine if any mutations of nucleobases were corrected or replaced along the way. Towards the end of the activity, you can begin tell the students what specific genes are activated by the miRNA molecules that you provided to them.

Comments: