Classroom Activities
Modeling Allele Frequency Changes Due to Selection Coefficients.
1. In a human population, the fitness advantage provided by the lactase persistence allele (L) can be represented by a selection coefficient (s). The selection coefficient s represents the relative fitness increase provided by the allele.
Suppose the initial frequency of the lactase persistence allele is 0.30 (or 30%). We assume this allele's frequency increases linearly over time due to positive selection, with a selection coefficient of s = 0.025. Using Equation 1,
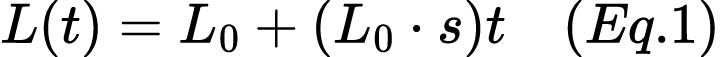
calculate the following:
- Verify the initial frequency of the lactase persistence allele at t = 0.
- Calculate the frequency of the lactase persistence allele at t=10 generations.
- Calculate the frequency of the lactase persistence allele at t=20 generations.
- Create a graph showing the change in frequency of the lactase persistence allele over 50 generations.
- Determine how many generations it will take for the frequency of the lactase persistence allele to reach 60%.
2. In a human population, the fitness advantage provided by the lactase persistence allele (L) can be represented by a selection coefficient (s). The selection coefficient represents the relative fitness increase provided by the allele. Suppose the initial frequency of the lactase persistence allele is 0.25 (or 25%). We assume this allele's frequency increases linearly over time due to positive selection, with a selection coefficient of s=0.02.
Using equation 1, calculate the following: Verify the initial frequency of the lactase persistence allele at t=0; calculate the frequency of the lactase persistence allele at t=15 generations; calculate the frequency of the lactase persistence allele at t=30 generations; create a graph showing the change in frequency of the lactase persistence allele over 60 generations; determine how many generations it will take for the frequency of the lactase persistence allele to reach 50%.
Modeling Allele Frequency Changes Due to Mutation Rates.
3. Let us consider a human population where the frequency of the lactase persistence allele is increasing due to both mutation and positive selection. The mutation rate for lactase persistence (u) is 0.001 mutations per generation, and the selection coefficient (s) representing the advantage of lactose tolerance is 0.02. Initially, 20% of the population is lactose tolerant.
We can model the frequency of the lactase persistence allele (L) in the population at time t (measured in generations) using a linear approximation over short periods using Equation 2:
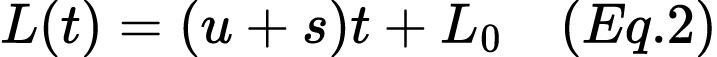
Calculate the following,
- Verify the initial frequency of the lactase persistence allele at t = 0.
- Calculate the frequency of the lactase persistence allele at t=10 generations.
- Calculate the frequency of the lactase persistence allele at t=20 generations.
- Create a graph showing the change in frequency of the lactase persistence allele over 50 generations.
- Determine how many generations it will take for the frequency of the lactase persistence allele to reach 60%.
- Compare the answer with problem one, which does not involve mutations.
4. Consider a human population where the frequency of an allele for disease resistance, denoted as L, increases due to both mutation and positive selection. The mutation rate for this allele (u) is 0.0008 mutations per generation, and the selection coefficient (s) representing the advantage of disease resistance is 0.025. Initially, 30% of the population has the disease resistance trait. We can model the frequency of the disease resistance allele L in the population at time t (measured in generations) using a linear approximation over short periods using Equation 2:
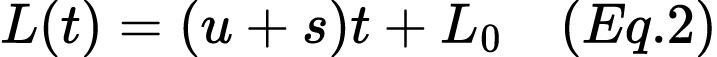
Please calculate the following:
- Verify the initial frequency of the disease-resistance allele at t=0.
- Calculate the frequency of the disease resistance allele at t=10t=10t=10 generations.
- Calculate the frequency of the disease resistance allele at t=20t=20t=20 generations.
- Create a graph showing the change in frequency of the disease resistance allele over 50 generations.
- Determine how many generations it will take for the frequency of the disease-resistance allele to reach 70%.
- Compare the answer with a scenario without mutations (i.e., u=0).
Modeling Allele Frequency Changes Due to selection coefficients and mutation rates.
5. Let us consider a human population where the frequency of the lactase persistence allele is increasing due to both mutation and positive selection. The mutation rate for lactase persistence (u) is 0.001 mutations per generation, and the selection coefficient (s) representing the advantage of lactose tolerance is 0.03. Initially, 25% of the population is lactose tolerant. We can model the frequency of the lactase persistence allele (L) in the population at time t (measured in generations) using a linear approximation over short periods using Equation 2.
Please calculate the following: verify the initial frequency of the lactase persistence allele at t=0; calculate the frequency of the lactase persistence allele at t=10 generations; calculate the frequency of the lactase persistence allele at t=20 generations; create a graph showing the change in frequency of the lactase persistence allele over 50 generations; determine how many generations it will take for the frequency of the lactase persistence allele to reach 60%; compare the answer with a problem that does not involve mutations.
6. Consider a human population where the frequency of the allele for lactase persistence (the ability to digest lactose), denoted as L, is increasing due to both mutation and positive selection. The mutation rate for lactase persistence (u) is 0.001 mutations per generation, and the selection coefficient (s) representing the advantage of lactose tolerance is 0.03. Initially, 25% of the population is lactose tolerant. We can model the frequency of the lactase persistence allele L in the population at time t (measured in generations) using a linear approximation over short periods with equation 2:
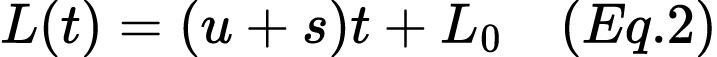
Please calculate the following:
- Verify the initial frequency of the lactase persistence allele at t=0.
- Calculate the frequency of the lactase persistence allele at t=10 generations.
- Calculate the frequency of the lactase persistence allele at t=20 generations.
- Create a graph showing the change in frequency of the lactase persistence allele over 50 generations.
- Determine how many generations it will take for the frequency of the lactase persistence allele to reach 60%.
- Compare the answer with a scenario without mutations (i.e., u=0).
Modeling Allele Frequency Changes Due to Genetic Drift and Randomness
7. In a population of 500 individuals, the initial frequency of the lactase persistence allele (L) is 0.2. Using the linear model for genetic drift, calculate the expected allele frequency after 20 generations. Assume that the random change in allele frequency per generation is ΔL=0.01. Use Equation 3:
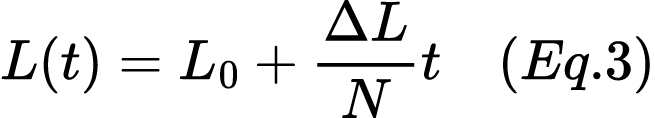
8. In a population of 500 individuals, the initial frequency of the lactase persistence allele (denoted as L) is 0.25. Using the linear model for genetic drift, calculate the expected allele frequency after 20 generations. Assume that the random change in allele frequency per generation is ΔL=0.008. Use Equation 3:
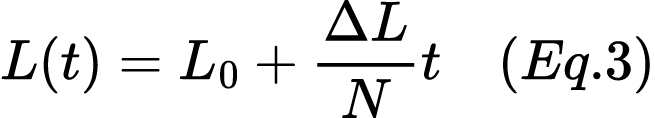
Modeling Allele Frequency Changes Due to Genetic Drift and Population Size
9. Consider two populations with different sizes: Population A has 200 individuals, and Population B has 1,000 individuals. The initial frequency of the lactase persistence allele (L) in both populations is 0.3. Using the linear model for genetic drift, calculate the expected allele frequency after 15 generations in both populations. Assume the random change in allele frequency per generation is ΔL=005. Use Equation 3:
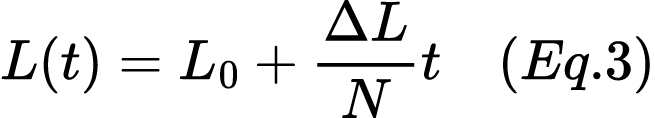
10. Consider two populations with different sizes: Population X has 300 individuals, and Population Y has 1,500 individuals. The initial frequency of the lactase persistence allele (denoted as L) in both populations is 0.35. Using the linear model for genetic drift, calculate the expected allele frequency after 20 generations in both populations. Assume the random change in allele frequency per generation is ΔL=0.004. Use Equation 3:
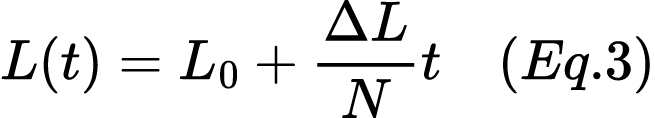
Modeling Allele Frequency Changes Due to Genetic Drifting and the Bottleneck Effect
11. A population of 1,000 individuals experiences a bottleneck event due to a natural disaster, reducing its size to 50 individuals. Before the bottleneck, the lactase persistence allele (L) frequency is 0.4. Calculate the expected allele frequency after ten generations using the linear model for the bottleneck effect. Assume the random change in allele frequency per generation due to genetic drift is ΔL=0.01. Use Equation 3:
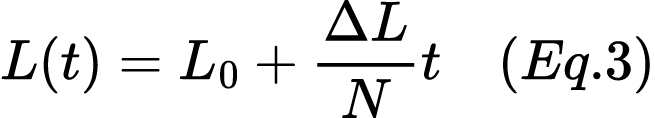
12. A population of 800 individuals experiences a bottleneck event due to a disease outbreak, reducing its size to 40 individuals. Before the bottleneck, the frequency of the lactase persistence allele (denoted as L) is 0.45. Calculate the expected allele frequency after ten generations using the linear model for the bottleneck effect. Assume the random change in allele frequency per generation due to genetic drift is ΔL=0.012. Use Equation 3:
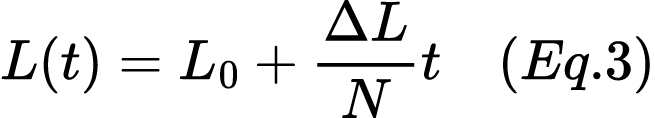
Please calculate the following:
- Verify the initial frequency of the lactase persistence allele at t=0t=0t=0.
- Calculate the expected frequency of the lactase persistence allele after 5 generations.
- Calculate the expected frequency of the lactase persistence allele after ten generations.
- Discuss how the bottleneck event might impact the population's genetic diversity over time.
Modeling Allele Frequency Changes Due to Genetic Drifting and the Founder Effect
13. A group of 20 individuals from a larger population migrates to an isolated island, forming a new population. The initial frequency of the lactase persistence allele (L) in this founder population is 0.6. Calculate the expected allele frequency after 15 generations, assuming the random change in allele frequency per generation is ΔL=0.02. Use Equation 3:
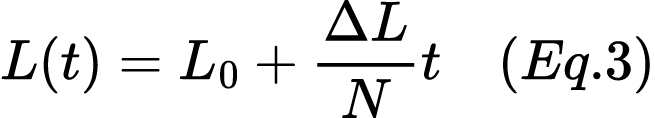
14. A group of 25 individuals from a larger human population migrates to a remote island, forming a new population. The lactase persistence allele's initial frequency (L) in this founder population is 0.55. Calculate the expected allele frequency after 15 generations, assuming the random change in allele frequency per generation is ΔL=0.015. Use Equation 3:
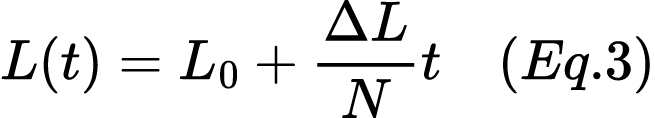
Please calculate the following:
- Verify the initial frequency of the lactase persistence allele at t=0.
- Calculate the expected frequency of the lactase persistence allele after 5 generations.
- Calculate the expected frequency of the lactase persistence allele after ten generations.
- Calculate the expected frequency of the lactase persistence allele after 15 generations.
- Discuss how the founder effect might impact the genetic diversity of the new population over time.
Modeling Allele Frequency Changes Due to selection coefficients and randomness.
15. A population of 500 individuals has an initial frequency of the lactase persistence allele (L) of 0.5. The allele has a selection coefficient of s=0.02, giving it a slight fitness advantage. Calculate the expected allele frequency after ten generations, assuming the random change in allele frequency per generation due to drift is ΔL=0.01. Use equation 4:
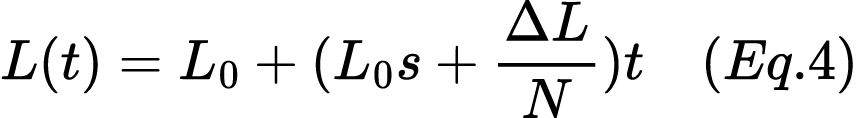
16. A population of 500 humans has an initial frequency of the lactase persistence allele (denoted as L) of 0.5. The allele has a selection coefficient of s=0.02, giving it a slight fitness advantage. Calculate the expected allele frequency after ten generations, assuming the random change in allele frequency per generation due to drift is ΔL=0.01. Use Equation 4:
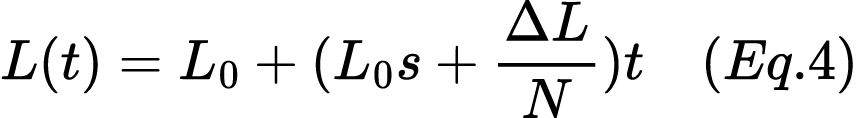
Please calculate the following:
- Verify the initial frequency of the lactase persistence allele at t=0t=0t=0.
- Calculate the expected frequency of the lactase persistence allele after 5 generations.
- Calculate the expected frequency of the lactase persistence allele after ten generations.
- Discuss the impact of genetic drift and selection on the allele frequency over the ten generations.
Modeling Allele Frequency Changes Due to selection coefficients and mutation rates.
17. A population of 1,000 individuals experiences a bottleneck event, reducing its size to 50 individuals. The initial frequency of the lactase persistence allele (L) is 0.3. This allele has a selection coefficient of s=0.02, providing a fitness advantage. The mutation rate introducing this allele into the population is u = 0.001. Calculate the expected allele frequency after 15 generations, accounting for the bottleneck effect, with the random change in allele frequency per generation due to drift represented by ΔL=0.01. Use Equation 5:
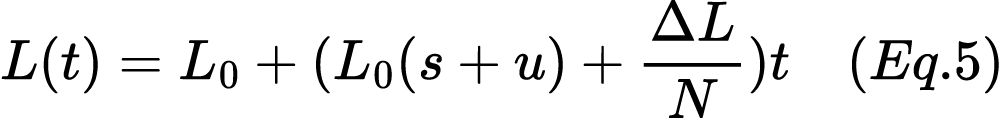
18. A population of 1,200 individuals experiences a bottleneck event due to a natural disaster, reducing its size to 60 individuals. The initial frequency of the lactase persistence allele (L) is 0.35. This allele has a selection coefficient of s=0.025, providing a fitness advantage. The mutation rate introducing this allele into the population is u=0.0012. Calculate the expected allele frequency after 15 generations, accounting for the bottleneck effect, with the random change in allele frequency per generation due to drift represented by ΔL=0.012. Use Equation 5:
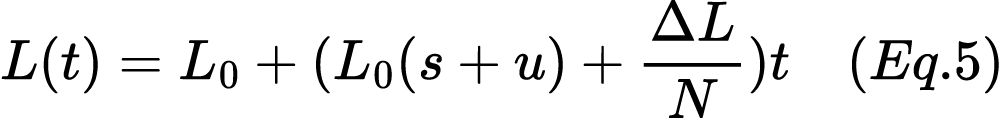
Please calculate the following:
- Verify the initial frequency of the lactase persistence allele at t=0t=0t=0.
- Calculate the expected frequency of the lactase persistence allele after 5 generations.
- Calculate the expected frequency of the lactase persistence allele after ten generations.
- Calculate the expected frequency of the lactase persistence allele after 15 generations.
- Discuss how the bottleneck event and mutation rate might impact the population's genetic diversity.

Comments: